This retrospective study was performed with the understanding and informed consent of the subjects. All of the samples used in this study are the property of the tissue collection of the Pathology Department of the University Hospital of Nice and are declared annually to the French Health Ministry. The procedures followed were approved by the institutional review board of the University Hospital of Nice. This study was conducted in accordance with the Declaration of Helsinki.
State-of-the-art convolutional neural networks
We want to assess the efficiency of the state-of-the-art CNNs to correctly categorize the four different subtypes of RCC neoplasms and also to define the most performing training configuration.
To answer these questions, six consolidated deep network models have been put into effect through TensorFlow framework: VGG1628, ResNet5029, ResNet10129, Inception Xception30,31, DenseNet12132, and ConvNeXt33. CNNs typically need a large amount of labeled data to learn good visual representations, while preparing large-scale labeled datasets is expensive and time-consuming, especially for medical image data9,34. Hence, to avoid, or at least to limit, this tedious data collection and annotation phase, some researchers take as compromise ImageNet-pretrained convolutional neural network to extract visual representations from a large set of different image types, the last training steps being performed on a reduced medical images database9,34.
On top of this consideration, each CNN we implemented was trained following three different learning paradigms: (i) training from scratch; (ii) transfer learning leveraging ImageNet as source domain; (iii) transfer learning leveraging a different histological dataset as source domain. In this latter experimental configuration, we exploited the pre-training on the Colorectal Cancer (CRC) classification task described in a recent study by Ponzio et al.9 to extract visual representations closer to our final target dataset, i.e. the RCC.
To obtain representative training and testing sets, in terms of inter-subjects and inter-class variability, we opted to randomly separate 54 patients for training and to leave 37 for testing our models, i.e. with a 60/40 ratio (see Table 4). The specimen (i.e. WSIs) selected as training set, were subsequently divided by a pathologist into regions of interest (ROIs) leveraging the so-called ROI-cropping procedure27,35, consisting in: (i) manually dividing each slide into ROIs that are homogeneous in terms of tissue content; (ii) manually annotating the ROIs, imposing a unique label to each tissue category; (iii) dividing the ROIs into a regular grid of tiles, that can be fed into the networks. Note that, through the above-mentioned procedure, the pathologist selected ROIs depicting several different tissue types, namely: four RCC subtypes (ccRCC, papRCC, chrRCC, ONCO) and a not-cancer super-class (including fiber, necrosis and normal renal parenchyma).
The tiles obtained through the ROI-cropping were subsequently divided into a training and a validation set with a 75–25% random split (see Fig. 2a), ensuring that regions coming from a single subject always belong to the same set. These sets were exploited in a threefold cross-validation fashion to find optimal hyper-parameters for the canonical CNNs as well as for our ExpertDT, as later described.
Accordingly with pathologist’s expertise17, the tile size has been set to \(1000 \times 1000\) with a downstream scaling to \(112 \times 112\). A second independent cohort of 37 RCC patients, never used during the training of the models, nor for the hyper-parameters optimization phase, were randomly selected to act as the test set for performance evaluation in terms of patient-level predictions.
As Fig. 2c suggests, for both the canonical CNNs and for our ExpertDT, the final classification at patient-level comes from a majority voting among the predicted tiles associated to the same subject.
For all the different CNN models exploited in the RCC subtyping task (VGG1628, ResNet50, ResNet10129, DenseNet12132, Inception Xception30,31, ConvNeXt33), and their corresponding training paradigms (training from scratch, transfer learning from ImageNet, transfer learning from CRC dataset36), we leveraged a grid search based on the KerasTuner package37 to look for the optimal configuration of the following hyper-parameters: the layer from which the fine-tuning starts (when transfer learning is employed), the learning rate and the optimizer type. Such optimization was done on a specific partition of the training set, and no patients from the test set have been considered.
In particular, we found the VGG16 model pre-trained on the ImageNet starting from the 11th layer as the optimal model. The learning rate was \(1\textrm{e}{-5}\) with Adam optimizer. For all the tested models, we leveraged the original network architecture described in the corresponding paper, and we set batch size equal to 128 images. All the models were trained for at most 150 epochs, leveraging an early stopping criterion based on the training loss (loss no longer decreasing for more than 20 epochs).
ExpertDeepTree’s training
As it can be gathered from Fig. 2b, the backbone of our ExpertDT consists of binary CNN classifiers (grey trapezoids in Fig. 2b) arranged in a tree-style architecture, which directly stems from the pathologist’s experience, and thus is responsible for the introduction of expert-based knowledge in our DL system.
Each binary CNN is individually trained on a reduced subset of the training dataset showing only the two labels of interest for the given binary classification task, artificially balanced via random under-sampling38. Specifically:
-
1.
the Root CNN learns the classification between tumor (T) and not-tumor (NT). The T class includes all the cancer subtypes, while the NT class is made up of tissue identified as not-cancerous by the pathologist (fiber, necrosis and normal renal parenchyma).
-
2.
the Node discriminates between the two super-labels \(pap+cc\) and \(chr+ONCO\), respectively obtained from the union of papRCC with ccRCC, and chrRCC with ONCO. The specific arrangement of the two super-classes stems from the pathologist’s expertise: it is easier to categorize between the union of ccRCC and papRCC versus the union of chrRCC and ONCO, with respect to any other super-class layout or with respect to a canonical 5 class staging. Moreover, is convenient to focus on peculiar differential diagnosis between ccRCC vs. papRCC and chrRCC vs. ONCO with respect to a task made up of more categories together. This last step is put into effect by the ExpertDT’s leaves.
-
3.
the Leaf1 categorizes chrRCC vs. ONCO.
-
4.
the Leaf2 classifies ccRCC vs. papRCC.
The optimal hyper-parameters configuration for all the CNNs backbone of ExpertDT was identified on the validation set in a twofold cross-validation fashion among non-overlapping groups of patients, following the same procedure as the one described in the previous subsection. We found again the VGG16 pre-trained on the ImageNet starting from the 11th layer as the optimal model. The learning rate was \(1\textrm{e}{-5}\), Adam was the optimizer, and all the models were trained for at most 150 epochs, leveraging an early stopping criterion based on the training loss (loss no longer decreasing for more than 20 epochs). Refer to Supplementary Fig. 1 for the classification performance of the single binary classifiers backbones of the proposed ExpertDT.
ExpertDeepTree’s testing
In our ExpertDT, each CNN model, identified in Fig. 2b by means of a grey trapezoid, is a binary classifier, whose two class predictions are represented as circles. Grey circles correspond to branch labels, while green circles to leaves. At the inference phase, a branch is a temporary label which leads the given testing crop \(x^*\) to the subsequent classification step. The whole classification process ends when \(x^*\) reaches a leaf, which corresponds to the final class associated with it. The final classification label, output of our ExpertDT, is provided at patient-level. Since the WSIs must be cropped into thousands of crops to be fed to the proposed architecture (see Fig. 2b on the left), the final decision at patient-level derives from a majority voting among the predicted crops associated to the same patient, and excluding those crops predicted as not-tumor (NT leaf in Fig. 2b). Note that, when a given WSI is fed to our system to be classified, the first preprocessing step is the background removal. The tiles recognised as background (see the transparent part in the WSIs reported in Fig. 2) are removed from the testing pipeline, and thus are not classified. The background removal has been carried out by simply defining an average value threshold of tile pixels to eliminate empty areas, namely where the tissue is almost absent. The corresponding threshold on the mean pixel value was empirically set to 210 on the training set.
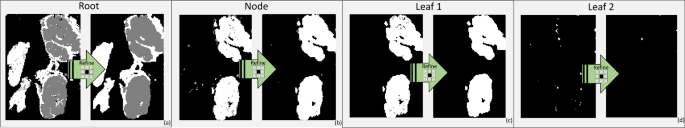
The Refine low-pass filtering effect
Downstream to each classification stage, we implemented the so-called Refine smoothing effect (see the symbol Refine in Fig. 2b). Such mechanism acts as a low-pass, denoising filter capable of relabelling isolated miss-classified tiles depending on the majority voting of the neighbourhood. It works as follows: for the generic testing crop \(x^*\) we define: (i) its corresponding prediction \(p^*\); (ii) its 9-connected neighbourhood \(9N^*\), including those crops that touch either one of the edges or the corners of \(x^*\) plus the pixel itself; (iii) the array of the predictions of the crops included in \(9N^*\), referred to as \(\vec {p^*}\).
To obtain the desired low-pass denoising effect, \(p^*\) is substituted with the value obtained by the majority voting among \(\vec {p^*}\). Note that, as previously mentioned, the background tiles are not fed to our model and hence not considered also in the Refine process. Figure 5 shows the effect of the different Refine phases implemented after each classification stage. As it can be gathered from the figure, the Refine is able to remove crops whose classification differs from its neighbourhood, which typically indicates a miss-classified crop.
Naive trees
The architectures of the NaiveDT versions derive from the two other possible permutations of the Node’s structure: \(pap+ONCO\) versus \(chr+cc\) or \(pap+chr\) versus \(ONCO+cc\). Thus, they are not related to any pathologist’s expertise. For both NaiveDT1 and NaiveDT2, the Root CNN learns the classification between tumor (T) and not-tumor (NT). This stage is the same as in ExpertDT. Conversely, the node classification is specific for the given NaiveDT versions: \(pap+ONCO\) versus \(chr+cc\) for NaiveDT1, \(pap+chr\) versus \(ONCO+cc\) for NaiveDT2. Lastly, Leaf1, and Leaf2 categorizations directly depends on the associated node: pap versus ONCO and chr versus cc for NaiveDT1; pap versus chr and ONCO versus cc for NaiveDT2.

Be the first to comment