From mathematical foundations to edge implementation
👨🏽💻 Github: thommaskevin/TinyML (github.com)
👷🏾 Linkedin: Thommas Kevin | LinkedIn
📽 Youtube: Thommas Kevin — YouTube
👨🏻🏫 Research group: Conecta.ai (ufrn.br)
SUMMARY
1 — Introduction to Long Short-Term Memory (LSTM)
1.1 — Forget Gate
1.2 —Input Gate and Candidate Memory
1.3 — Output Gate
1.4 — Backward Pass
2 — TinyML Implementation
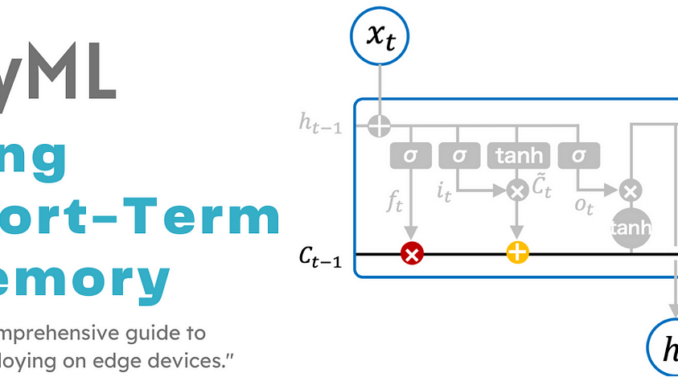
The neural network architecture for a Long Short-Term Memory (LSTM) block, shown in Figure 4, demonstrates that the LSTM network extends the memory of Recurrent Neural Networks (RNNs) and can selectively remember or forget information through structures called cell states and three gates. Therefore, in addition to a hidden state in RNNs, an LSTM block typically has four additional layers. These layers are the cell state (Ct), an input gate (it), an output gate (Ot), and a forget gate (ft). Each layer interacts with the others in a unique way to generate information from the training data.
In the forget gate, a sigmoid activation function is utilized. For the input and output gates, a combination of the sigmoid and hyperbolic tangent (tanh) functions is employed to provide the necessary information to the cell state. The information generated by these blocks flows through the cell state from one block to another, maintaining the chain of repeating components characteristic of the LSTM neural network. Details about the cell state and each layer are provided in subsequent sections.
The sigmoid activation function is defined as:
The hyperbolic tangent activation function is defined as:
1.1 — Forget Gate
The Forget Gate (F_t) decides the type of information that should be discarded or retained in the cell state. This process is implemented using a sigmoid activation function. The sigmoid activation function outputs values between 0 and 1 based on the weighted input (W_f p_t), the previous hidden state (h_{t-1}), and a bias (b_f).
Here, σ is the sigmoid activation function, W_f and b_f are the weight matrix and bias vector, which are learned from the input training data.
The function takes the previous hidden state (h_{t-1}) at time (t-1) and the current input (p_t) at time (t) to calculate the components that control the cell state and hidden state of the layer. The results range from 0 to 1, where 1 represents “completely hold this” and 0 represents “completely throw this away”.
1.2 — Input Gate and Candidate Memory
The Input Gate (I_t) controls what new information will be added to the cell state from the current input. This gate also plays a role in protecting the memory contents from perturbation by irrelevant input (Figure 3).
A sigmoid activation function is used to generate the input values and converts information between 0 and 1. Mathematically, the input gate is:
where (W_i) and (b_i) are the weight matrix and bias vector, (p_t) is the current input, and (h_{t-1}) is the previous hidden state. Similar to the forget gate, the parameters in the input gate are learned from the input training data. At each time step, with the new information (p_t), we can compute a candidate cell state.
Next, a vector of new candidate values, (\tilde{C}_t), is created. The computation of the new candidate is similar to that of the forget gate but uses a hyperbolic tangent (tanh) activation function with a value range of (-1, 1). This leads to the following equation (5) at time (t):
In the next step, the values of the input gate and the cell candidate are combined to create and update the cell state as given in equation (5). The linear combination of the input gate and forget gate is used for updating the previous cell state (C_{t-1}) into the current cell state (C_t). The input gate (i_t) determines how much new data should be incorporated via the candidate (tilde{C}_t), while the forget gate (f_t) determines how much of the old memory cell content (C_{t-1}) should be retained. Using pointwise multiplication (Hadamard product), we arrive at the following updated equation:
1.3 — Output Gate
The Output Gate (O_t) controls which information to reveal from the updated cell state (C_t) to the output in a single time step. In other words, the output gate determines the value of the next hidden state at each time step. As depicted in Figure 4, the hidden state comprises information from previous inputs. Moreover, the calculated value of the hidden state for the given time step is used for the prediction (\hat{y_t} = softmax(W_y h_t + b_y)). Here, softmax is a nonlinear activation function.
The output gate is defined as:
- σ is the sigmoid function,
- Wo is the weight matrix for the output gate,
- bo is the bias vector for the output gate,
- pt is the current input,
- h_t−1 is the previous hidden state.
The next hidden state (h_t) is determined by applying the tanh activation function to the updated cell state (C_t) and then multiplying it by the output gate value (O_t):
Here, ⊙ denotes element-wise multiplication (Hadamard product). This process ensures that only relevant information from the cell state is passed to the next hidden state and ultimately to the output.
The prediction (\hat{y_t}) is then calculated as:
First, the previous hidden state (h_{t-1}) and the current input (p_t) are passed through a sigmoid function to determine the output gate value (O_t). Then, the updated cell state (C_t) is generated using the tanh function. Finally, the tanh output is multiplied by the sigmoid output to determine the information carried by the hidden state (h_t). The output of the output gate is an updated hidden state, used for prediction at time step (t).
The aim of this gate is to separate the updated cell state (which contains a lot of information not necessarily required in the hidden state) from the hidden state. The updated cell state (C_t) is critical as it influences the hidden state used in all gates of an LSTM block. The output gate assesses which parts of the cell state (C_t) are presented in the hidden state (h_t). The new cell and hidden states are then passed to the next time step (Figure 4).
1.4 — Backward Pass
The LSTM network generates an output (\hat{y_t}) at each time step that is used to train the network via gradient descent. During the backward pass, the network parameters are updated at each epoch (iteration). The main difference between the back-propagation algorithms of RNN and LSTM networks is a minor modification. The error term at each time step is calculated as (E_t = -y_t log(\hat{y_t})). Similar to RNNs, the total error is the sum of the errors from all time steps:
The gradient of the error with respect to the weights at each time step is calculated, and then the sum of the gradients over all time steps is obtained:
The predicted value (\hat{y_t}) is a function of the hidden state:
The hidden state (h_t) is a function of the cell state:
Both of these functions are subject to the chain rule. Hence, the derivatives of the individual error terms with respect to the network parameters are:
For the overall error gradient using the chain rule of differentiation, we get:
The previous equation shows that the gradient involves the chain rule of in LSTM training using the backpropagation algorithm, while the gradient equation involves the chain rule for a basic RNN. Therefore, the Jacobian matrix for the cell state in an LSTM is:
With this example you can implement the machine learning algorithm in ESP32, Arduino, Arduino Portenta H7 with Vision Shield, Raspberry and other different microcontrollers or IoT devices.
2.0 — Install the libraries listed in the requirements.txt file
!pip install -r requirements.txt
2.1 — Importing libraries
from sklearn.model_selection import train_test_split
from eloquent_tensorflow import convert_model
import tensorflow as tf
from tensorflow.keras import layers
import pandas as pd
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
from tensorflow.keras.utils import plot_modelimport warnings
warnings.filterwarnings('ignore')
2.2 — Load Dataset
The “Vehicle Attributes and Emissions Dataset” contains comprehensive information on various vehicles manufactured in the year 2000. It includes details such as make, model, vehicle class, engine size, cylinder count, transmission type, and fuel type. Additionally, the dataset provides ranges for fuel consumption and CO2 emissions, offering insights into the environmental impact of each vehicle. The dataset encompasses a wide range of vehicle types, from compact to mid-size, and includes both conventional and high-performance models. With this information, analysts and researchers can study trends in vehicle characteristics, fuel efficiency, and emissions . This dataset serves as a valuable resource for understanding the automotive landscape and informing discussions on environmental sustainability and transportation policies.
link: https://www.kaggle.com/datasets/krupadharamshi/fuelconsumption/data
df = pd.read_csv('./data/FuelConsumption.csv')
df.head()
df.info()
df.describe()
2.3 — Clean Data
# 1. Removing rows with missing values
df.dropna(inplace=True)
# 2. Removing duplicates if any
df.drop_duplicates(inplace=True)
# Display the dataframe after cleaning
df.describe()
2.4 — Exploratory Data Analysis
sns.pairplot(df[['ENGINE SIZE','CYLINDERS','FUEL CONSUMPTION','COEMISSIONS ']])
plt.savefig('.\\figures\\pairplot.png', dpi=300, bbox_inches='tight')
corr = df[['ENGINE SIZE','CYLINDERS','FUEL CONSUMPTION','COEMISSIONS ']].corr('spearman')
# Adjusting the size of the figure
plt.figure(figsize=(18,10))
# Your existing code for generating the heatmap
heatmap = sns.heatmap(corr, xticklabels=corr.columns, yticklabels=corr.columns, cmap='coolwarm')
# Adding values to the heatmap
for i in range(len(corr.columns)):
for j in range(len(corr.columns)):
plt.text(j + 0.5, i + 0.5, f"{corr.iloc[i, j]:.2f}", ha='center', va='center', color='black', fontsize=18)
plt.xticks(fontsize=20, rotation=45)
plt.yticks(fontsize=20, rotation=0)
cbar = heatmap.collections[0].colorbar
cbar.ax.tick_params(labelsize=20)
plt.savefig('.\\figures\\heatmap.png', dpi=300, bbox_inches='tight')
# Display the heatmap
plt.show()
2.5 — Split into training and test data
X=df[['ENGINE SIZE','CYLINDERS', 'COEMISSIONS ']]
y=df[['FUEL CONSUMPTION']]
# Split the data into training and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
2.6 — Create the Long Short-Term Memory (LSTM)
def instantiate_lstm_for_regression(input_shape, output_shape):
model = tf.keras.Sequential()
model.add(layers.Input(shape=input_shape, batch_size=1))
model.add(layers.LSTM(12, unroll=False, return_sequences=True))
model.add(layers.LSTM(12, unroll=False))
model.add(layers.Dense(32, activation='relu'))
model.add(layers.Dense(output_shape, activation='linear'))
model.compile(optimizer='adamax', loss='mae', metrics=['mae'])
return model
model = instantiate_lstm_for_regression(input_shape = (3,1), output_shape = 1)
print(model.summary())
plot_model(model, to_file='./figures/model.png')
2.7 — Train the model
history = model.fit(X_train, y_train, validation_split=0.2, epochs=300)
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(1, len(loss) + 1)
plt.plot(epochs, loss, 'r.', label='Training loss')
plt.plot(epochs, val_loss, 'y', label='Validation loss')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.grid()
plt.legend()
plt.savefig('.\\figures\\history_traing.png', dpi=300, bbox_inches='tight')
plt.show()
2.8 — Model evaluation
y_train_pred = model.predict(X_train)
y_test_pred = model.predict(X_test)
# Calculate residuals
train_residuals = y_train.values.reshape(1,-1).tolist()[0] - y_train_pred
# Calculate mean and standard deviation of residuals
train_residuals_mean = np.mean(train_residuals)
train_residuals_std = np.std(train_residuals)
# Calculate residuals
test_residuals = y_test.values.reshape(1,-1).tolist()[0] - y_test_pred
# Calculate mean and standard deviation of residuals
test_residuals_mean = np.mean(test_residuals)
test_residuals_std = np.std(test_residuals)
# Plot residuals
plt.figure(figsize=(10, 5))plt.subplot(1, 2, 1)
plt.scatter(y_train_pred, train_residuals, c='blue', marker='o', label=f'Training data')
plt.axhline(y=0, color='r', linestyle='-')
plt.axhline(y=train_residuals_mean, color='k', linestyle='--', label=f'Mean: {train_residuals_mean:.3f}')
plt.axhline(y=train_residuals_mean + 2 * train_residuals_std, color='g', linestyle='--', label=f'+2 Std Dev: {2*train_residuals_std:.2f}')
plt.axhline(y=train_residuals_mean - 2 * train_residuals_std, color='g', linestyle='--', label=f'-2 Std Dev: {-2*train_residuals_std:.2f}')
plt.xlabel('Predicted values')
plt.ylabel('Residuals')
plt.title('Residuals vs Predicted values (Training data)')
plt.legend(loc='upper left')
plt.grid(True)
plt.subplot(1, 2, 2)
plt.scatter(y_test_pred, test_residuals, c='green', marker='s', label=f'Test data')
plt.axhline(y=0, color='r', linestyle='-')
plt.axhline(y=test_residuals_mean, color='k', linestyle='--', label=f'Mean: {test_residuals_mean:.3f}')
plt.axhline(y=test_residuals_mean + 2 * test_residuals_std, color='g', linestyle='--', label=f'+2 Std Dev: {2*test_residuals_std:.2f}')
plt.axhline(y=test_residuals_mean - 2 * test_residuals_std, color='g', linestyle='--', label=f'-2 Std Dev: {-2*test_residuals_std:.2f}')
plt.xlabel('Predicted values')
plt.ylabel('Residuals')
plt.title('Residuals vs Predicted values (Test data)')
plt.legend(loc='upper left')
plt.grid(True)
plt.tight_layout()
plt.show()
# Check for normality
plt.figure(figsize=(10, 5))
plt.subplot(1, 2, 1)
plt.hist(train_residuals, bins=20, color='blue', alpha=0.6)
plt.title('Histogram of Residuals (Training data)')
plt.xlabel('Residuals')
plt.ylabel('Frequency')
plt.axvline(x=train_residuals_mean, color='k', linestyle='--', label=f'Mean: {train_residuals_mean:.3f}')
plt.axvline(x=train_residuals_mean + 2 * train_residuals_std, color='g', linestyle='--', label=f'+2 Std Dev: {2*train_residuals_std:.3f}')
plt.axvline(x=train_residuals_mean - 2 * train_residuals_std, color='g', linestyle='--', label=f'-2 Std Dev: {-2*train_residuals_std:.3f}')
plt.legend(loc='upper right')
plt.grid(True)
plt.subplot(1, 2, 2)
plt.hist(test_residuals, bins=20, color='green', alpha=0.6)
plt.title('Histogram of Residuals (Test data)')
plt.xlabel('Residuals')
plt.ylabel('Frequency')
plt.axvline(x=test_residuals_mean, color='k', linestyle='--', label=f'Mean: {test_residuals_mean:.3f}')
plt.axvline(x=test_residuals_mean + 2 * test_residuals_std, color='g', linestyle='--', label=f'+2 Std Dev: {2*test_residuals_std:.3f}')
plt.axvline(x=test_residuals_mean - 2 * test_residuals_std, color='g', linestyle='--', label=f'-2 Std Dev: {-2*test_residuals_std:.3f}')
plt.legend(loc='upper right')
plt.grid(True)
plt.tight_layout()
plt.show()
2.8.1 — Evaluating the model with train data
plt.plot(y_train, label="original")
plt.plot(y_train_pred, label="predicted")
plt.legend(loc='best',fancybox=True, shadow=True)
plt.grid()
plt.show()
2.8.2 — Evaluating the model with test data
plt.plot(y_test.values, label="original")
plt.plot(y_test_pred, label="predicted")
plt.legend(loc='best',fancybox=True, shadow=True)
plt.grid()
2.9 — Obtaining the model to be implemented in the microcontroller
# Example usage of convert_model
code = convert_model(model)
2.10 — Saves the template in a .h file
with open('./LSTM/model.h', 'w') as file:
file.write(code)
2.11 — Deploy Model
Import the libraries into your Arduino sketch under libraries -> ESP32 -> EloquentTinyML-main.zip and tflm_esp32–2.0.0.zip
2.11.1 — Complete Arduino Sketch
#include
// replace with your own model
#include "model.h"
// include the runtime specific for your board
// either tflm_esp32 or tflm_cortexm
#include
// now you can include the eloquent tinyml wrapper
#include // this is trial-and-error process
// when developing a new model, start with a high value
// (e.g. 10000), then decrease until the model stops
// working as expected
#define ARENA_SIZE 30000
Eloquent::TF::Sequential tf;
float X_1[3] = {4.6, 8., 304.};
float X_2[3] = {2., 4., 216.};
void predictSample(float *input, float expectedOutput) {
while (!tf.begin(tfModel).isOk())
{
Serial.println(tf.exception.toString());
delay(1000);
}
// classify class 0
if (!tf.predict(input).isOk()) {
Serial.println(tf.exception.toString());
return;
}
Serial.print("Expcted = ");
Serial.print(expectedOutput);
Serial.print(", predicted = ");
Serial.println(tf.outputs[0]);
}
void setup()
{
Serial.begin(115200);
delay(3000);
Serial.println("__TENSORFLOW LSTM__");
// configure input/output
// (not mandatory if you generated the .h model
// using the eloquent_tensorflow Python package)
tf.setNumInputs(TF_NUM_INPUTS);
tf.setNumOutputs(TF_NUM_OUTPUTS);
registerNetworkOps(tf);
}
void loop()
{
/**
* Run prediction
*/
predictSample(X_1, 17.76);
delay(2000);
predictSample(X_2, 11.44);
delay(2000);
}
2.12 — Results
Full project in: TinyML/18_LSTM at main · thommaskevin/TinyML (github.com)
If you like it, consider buying my coffee ☕️💰 (Bitcoin)
code: bc1qzydjy4m9yhmjjrkgtrzhsgmkq79qenvcvc7qzn

Be the first to comment