Aggregation Tool for Genomic Concept
Current applications of machine learning to mutation data are generally limited to an aggregation of hand-crafted features. Our model differs in that attention is first given to individual instances before aggregation, and if desired an end-to-end model is possible allowing for instance features to be learned during training (known as representation learning). Hand-crafted feature engineering may be most efficient when the representation of an instance is well/completely understood and aggregation of information over a set of instances is done via a static function such as sum and mean. Representation learning at the level of the instance can extract features specific to a given machine learning task and, in scenarios with a very large possible set of genomic measures, can be combined with trainable attention mechanisms that can help with model explainability. To allow a model to extract its own features, decisions must be made on how the raw data will be presented to the model and how the model will encode the data. We consider the outcome of this process to be a genomic concept and essential to extracting relevant features. Somatic mutations are often reported in a Variant Call Format or Mutation Annotation Format (MAF), and a genomic concept can be constructed for any measurement in these files. The concept can be as simple as an embedding matrix (for example, our position encoder), or it can be as complex as convolutional layers for the flanking nucleotide sequences along with the reference and alteration in both the forward and reverse directions (our sequence encoder; Fig. 1).
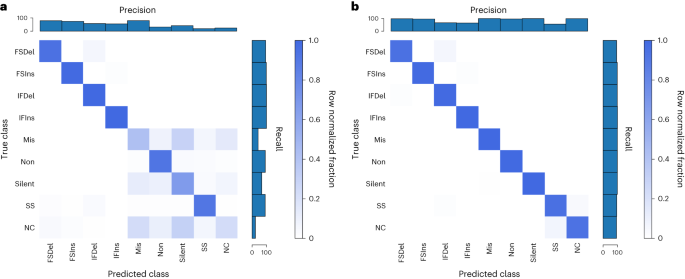
To confirm that the encoders we developed were valid, we performed positive controls, using the unique mutation calls from The Cancer Genome Atlas (TCGA) multi-centre mutation calling in multiple cancer (MC3) public MAF. Our sequence encoder was found to be a faithful representation of a variant, learning the 96 contexts and an outgroup with near-perfect accuracy, and our embedding strategy was able to perform a data compression without any information loss (Supplementary Fig. 1). To confirm that our sequence encoder could effectively utilize strand information, we asked a more difficult question: whether it could classify variants according to their consequence as provided by the MC3 MAF, specifically the consequence/variant classification of frameshift deletion, frameshift insertion, in-frame deletion, in-frame insertion, missense, nonsense, silent, splice site and noncoding (5′ untranslated region, 3′ untranslated region, intron). This problem requires learning all 64 codons in 6 different reading frames, and importantly the strand the variant falls on affects the label. We first asked how well the model could do without providing a reading frame, and while the model was able to learn insertions and deletions (InDels) and splice sites, it was unable to distinguish noncoding mutations from the other classes (as would be expected) and did the best it could at associating codons with a consequence (Fig. 2a). When provided, the reading frame in the form of strand and coding sequence position modulo 3 (noncoding variants were represented by a zero vector), the sequence concept was now able to correctly classify missense versus nonsense versus silent mutations (Fig. 2b), indicating that the modelling approach is able to learn which strand a feature was on and correctly associate the relevant codons with consequence.
a,b, Our sequence encoder can learn variant consequence as defined by variant effect predictor without (a) and clearly better with (b) reading frame information. FSDel, frameshift deletion; FSIns, frameshift insertion; IFDel, inframe deletion; IFIns, inframe insertion; Mis, missense; Non, nonsense; SS, splice site; NC, noncoding. All four plots show row normalized confusion matrices.
Our implementation of MIL is motivated by ref. 18 (Supplementary Fig. 2), with some important modifications for the nature of somatic mutation data. In image analysis the aggregation function is often a weighted average, but whereas the number of tiles is unrelated to the label for an image the number of mutations may provide information about a tumour sample. Using simulated data, we explored various MIL implementations along with traditional machine learning approaches and found our attention-based MIL with a weighted sum performed well (Supplementary Figs. 3–5). For a weighted sum to be meaningful, the instance features must be activated, and this results in potentially large values on the graph. To account for this, we perform a log of the aggregation on graph. We also developed dropout for MIL, wherein a random subset of instances is given to the model each gradient update, but then all instances are used during evaluation. This can allow for training with large samples and also helps with overfitting as the samples are altered every batch. To improve model explainability, we designed the model for multi-headed attention, where each attention head can be viewed as class-specific attention when the number of attention heads matches the number of classes. The model is implemented in TensorFlow with ragged tensors and is easily extensible to other data types, and we refer to the resulting tool as Aggregation Tool for Genomic Concepts (ATGC).
Cancer type classification
A readily available label in cancer datasets is the cancer type, and this task can have practical importance for when the tumour of origin for a metastatic cancer cannot be determined19. The types of mutation of a cancer are influenced by the mutational processes of its aetiology, while the genomic distribution of its mutations is influenced by histology of origin, and these features of somatic mutations have been shown to be capable of classifying cancers17,20,21,22. We were interested in seeing how a model that calculates attention or learns it owns features compared to established approaches. For this task we used the TCGA MC3 public mutation calls, which are exome based. To understand the baseline performance on this data using current approaches, we used two common hand-crafted features: the 96 single base substitutions (SBSs) contexts (and a 97th outgroup so that no data were discarded) and an approximately 1 Mb binning of the genome. For each manual feature, we ran a logistic regression, random forest, neural net and our MIL model. We also ran our model with 6 base pair (bp) windows of the sequences and explored gene as an input.
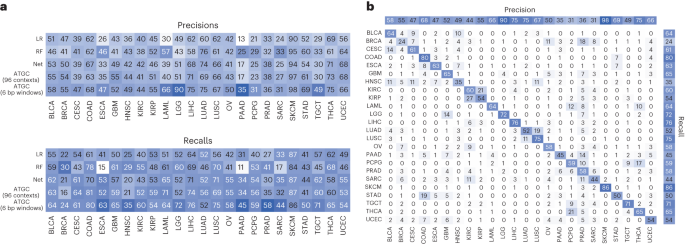
Table 1 shows the results of test folds from fivefold cross validation for the different models and the different inputs for the TCGA project codes (a 24-class problem). For each input our model outperformed current approaches, and the novel input of base-pair windows showed a significant benefit (~14% increase over the best standard model). Notably, our model showed a benefit even when using an identical encoding to the other models (96 contexts), suggesting the attention alone can improve model performance to some degree. To validate these results, we investigated the performance of the 96 contexts in whole genome sequencing and again saw a benefit with our model (86% accuracy compared to 81%; Supplementary Table 1). For additional validation we also ran the models classifying the MC3 samples according to their National Cancer Institute thesaurus (NCIt) codes and again saw a benefit with our model for every input (a 27-class problem; Supplementary Table 2).
To investigate the performance differences, we looked at classification performance in terms of precision and recall for each model stratified by tumour type (Fig. 3a). Our model, which takes 6 bp windows as input, showed significant improvement in oesophageal carcinoma, lower-grade glioma, pancreatic adenocarcinoma and testicular germ cell tumours. The improvement seen in lower-grade glioma is almost certainly due to identifying IDH1 mutations via a mapping of local sequence context of that specific hotspot. We also investigated the predictions of the best performing model (ATGC with 6 bp windows) with a confusion matrix (Fig. 3b) and observed that cancers of similar histologic origin were often mistaken for each other. For the corresponding analyses with gene as input, see Extended Data Fig. 1.
a, Precisions and recalls for the models using the 96 contexts and ATGC with the 6 bp windows. b, Confusion matrix for ATGC and the 6 bp windows. All numbers are shown as percentages. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; ESCA, oesophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukaemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; SARC, sarcoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumours; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma.
When investigating how the model is interpreting genomic variant data, we can examine both the representation of a variant produced by the model and what degree of attention the model is assigning a variant. To illustrate these two concepts, we show a heat map of the learned variant representation vectors for several cancer types with known aetiologies (Fig. 4). Unsupervised K-means clustering was used to group the instances within each tumour type, revealing a rich feature space for the learned variant representations and clear clusters for each cancer type. We next explored how class-specific attention related to this instance feature space and observed a spectrum of attention levels. In Fig. 4 we see that for skin cutaneous melanoma (SKCM) six clusters did for the most part produce clusters which were either high or low in attention, while in lung adenocarcinoma (LUAD) and colon adenocarcinoma (COAD) we see clusters that contain a bimodal distribution of attention.
The feature vectors for the instances of a single test fold for SKCM, LUAD and COAD samples were clustered with K-means into 6 clusters and ordered by their median class-specific attention (2,000 instances sampled at random per cancer shown in the heat maps). Violin plots of the attention values for each cluster are also shown along with sequence logos of the instances for the four clusters with highest median attention.
To investigate what sequences were present in each of these clusters, we generated sequence logos. For SKCM the highest attention cluster was composed of a specific doublet base substitution characteristic of ultraviolet radiation, with the next highest attention cluster comprising a very specific 5′ nucleotide, reference (ref), alternative (alt) and 3′ nucleotide also characteristic of ultraviolet radiation16. For LUAD the highest attention cluster contained sequences characteristic of tobacco smoking, while in COAD the highest attention cluster was a specific deletion occurring at a homopolymer, which is characteristic of cancers deficient in mismatch repair and is a known signature of this cancer type16.
While in Fig. 4 we used clustering to identify groups of mutations within a cancer type which may be of interest, we instead could have simply sorted all instances by attention. To explore this possibility, we took the highest attention instances (top 5%) of each attention head for SBSs and InDels and looked at the bits of information contained in the logos (Supplementary Fig. 6). For SBS mutations most of the information gain occurs at the alteration and flanking nucleotides; however, for most heads there is still information two, three or more nucleotides away from the alteration. Given that a head of attention may identify several distinct motifs as important, the high attention instances for each head may not be homogeneous, and as a result these bits represent a lower bound of the information gain. For InDels there is significant information three or four nucleotides away from the alteration, and these sequences are often mononucleotide repeats. When performing a similar analysis for our gene encoder, we noticed that a small number of genes appear to be given high attention in each head, and clear groupings were present in the embedding matrix, with a small cluster enriched in cancer-associated genes (Supplementary Fig. 7).
Through the attention mechanism we also investigated what our model learned about genomic location. When looking at the attention values for the 1 Mb bins, the different heads of attention appeared to be giving attention to the same bins, so we calculated the z-scores for each head and averaged across heads. We also noticed the values appeared consistent across folds of the data, so we also averaged over the five data folds. Figure 5a shows the averaged attention values for the bins across the genome along with an embedding matrix from one of data folds. Specific bins are clearly being either upweighted or downweighted. Investigation of bins with low attention scores revealed that they contained very little data, which caused us to wonder whether attention simply correlated with amount of data in each bin. There is initially a strong correlation between attention and number of mutations in a bin (Fig. 5b), but once bins contain a certain amount of data the relationship disappears. Given that this is exomic data, we suspected the bins with the highest attention contained genes important for cancer classification, so we calculated average gene attention z-scores with our model that used gene as input and matched genes with their corresponding bin (averaging when a gene was split across bins). There is nearly an order of magnitude more genes than bins, so most bins contain multiple genes, and the same attention will be assigned to all genes in the bin. For the genes with the highest attention there is a corresponding bin also being given high attention (Fig. 5c, top right corner), but there are also many genes that are passengers in those bins and mistakenly being given attention (Fig. 5c, top left corner). This likely explains why using genes as input outperformed models using position bins as input.
a, Attention z-scores for each 1 Mb bin were calculated across all 24 attention heads and averaged across 5 folds. The embedding matrix for a single fold is shown below with the starts of each chromosome shown. The z-scores between −1 and 1 are shaded. b, Attention z-scores for each 1 Mb bin plotted against the number of mutations falling in each bin. c, Attention z-scores for each 1 Mb bin plotted against the attention z-scores for the corresponding genes in those bins from a model with gene as input.
Microsatellite instability
Characterized by deficiencies in the mismatch repair proteins (MLH1, MSH2, MSH6, PMS2), microsatellite unstable tumours accumulate InDels at microsatellites due to polymerase slippage. As with tumour classification, current bioinformatic approaches to predicting microsatellite instability (MSI) status rely on manually featurizing variants. For example, the Microsatellite Analysis for Normal Tumor InStability (MANTIS) tool uses a binary alignment map file to calculate the average difference between lengths of the reference and alternative alleles at predefined loci known a priori to be important23. Similarly, the MSIpred tool calculates a 22-feature vector using information from a MAF file and loci again known to be important, specifically simple repeat sequences24. Both of these approaches are valid and produce accurate predictions, but they rely on knowing the nature of the problem.
Because mutations characteristic of MSI occur at many different repeat sites, and because the repeats have a distinctive sequence, we chose to use our sequence concept for this problem. For the data we opted to use the controlled TCGA MC3 MAF rather than the public MAF as the public MAF excludes most variants in non-exonic regions and most simple repeats fall in these regions. The TCGA has ground truth labels as defined by PCR assay for some tumour types, and we were able to obtain labels for uterine corpus endometrial carcinoma (494), stomach adenocarcinoma (437), COAD (365), rectum adenocarcinoma (126), oesophageal carcinoma (87) and uterine carcinosarcoma (56) tumour samples. For the sequence concept we went out to 20 nucleotides for each component (5′, 3′, ref and alt) to allow the model to potentially capture long repetitive sequences.
Although we did not provide the model information about cancer type or perform any sample weighting, our model showed similar performance across cancer types (Extended Data Fig. 2a). We believe MANTIS and MSIpred are considered state of the art when it comes to MSI classification performance, and as can be seen in Fig. 6a our model slightly outperforms them despite not being given information about simple repeats. When comparing our predictions to MANTIS, both models were often similarly confident in their predictions (Extended Data Fig. 2b); however, there are cases where MANTIS predicts the correct label but our model does not, or our model predicts the correct label but MANTIS does not, suggesting that the best MSI predictor may be one that incorporates predictions from multiple models. There are a few cases where the sample is labelled MSI high by the PCR, but both MANTIS and our model are confident the sample is MSI low, perhaps indicating the PCR label may not be 100% specific (or alternatively indicates an issue with the binary alignment map files for these samples).
a, Average precision recall curves over nine stratified K-fold splits for MSIpred and ATGC, precision recall curve for MANTIS for samples which had a MANTIS score. b, Probability density plots of the output of the attention layer stratified by whether a variant was considered to be in a simple repeat from the UCSC genome annotations. c, Features of 2,000 random instances from a single test fold clustered with K-means and the corresponding sequence logos for the 4 clusters with highest median attention.
To classify whether a variant falls in a repeat region or not, MSIpred relies on a table generated by the University of California, Santa Cruz (UCSC, http://hgdownload.cse.ucsc.edu/goldenPath/hg38/database/simpleRepeat.txt.gz). Using this file, we labelled variants according to whether they fell in these regions, and as seen in Fig. 6b variants which occurred in a simple repeat region were much more likely to receive high attention than variants that did not. When clustering the instances of these samples, we observed two clear clusters that are receiving high attention: a smaller cluster characterized by SBS mutations in almost exclusively intergenic regions with almost 33% labelled as a simple repeat and a larger cluster characterized by deletions in genic but noncoding regions. The sequence logo of the intergenic cluster did not reveal a specific sequence, while the logo for the deletion cluster revealed that the model is giving attention to deletions at a mononucleotide T repeat.




Be the first to comment